In 2020, researchers Jian Sun, Xiaoqin Huang et al, in the Laboratory of Immunology at the National Eye Institute, confirmed in the lab what has been going on in human ophthalmology in recent years, but with a twist. Their deep learning model enabled identification of retinal disease stages with noteworthy accuracy. What marked their work as distinct is they were the first to successfully apply artificial intelligence to a dataset of 1500 Phoenix MICRON® retinal images of a mouse model of uveitis. Their findings hold exciting promise for developing tools for both experimental models as well as for clinical practice.
Uveitis describes a set of conditions characterized by intraocular inflammation. It is believed to account for 10% of legal blindness in the USA. Clinical assessment is challenging and complicated therefore evaluation of retinal change is prone to inter and intra-observer variability. Accurate detection is key to tailoring appropriate medical treatments that carry risks of significant adverse ocular and systemic side effects.
In clinical practice, artificial intelligence (AI) identification of retinal disease has been successfully used in diabetic retinopathy, glaucoma, age-related macular degeneration and retinopathy of prematurity with accuracy comparable to that of experienced ophthalmologists.
Benefits include 1) consistency between intra and inter observer analysis, 2) learning accuracy of the algorithm and 3) cost-effectiveness compared with extensive training of human experts. While this describes the findings in the medical community, it also holds true in the world of animal research.
Experimental autoimmune uveitis (EAU) is often used as the animal model for uveitis as it shares essential features with the human form. EAU is a challenging disease to successfully and reproducibly classify both clinically and experimentally. Sun and colleagues applied their convolutional neural network (CNN) model to the identification of 3 classes of disease severity (normal, trace and disease) in EAU with comparable, if not slightly better precision than human graders.
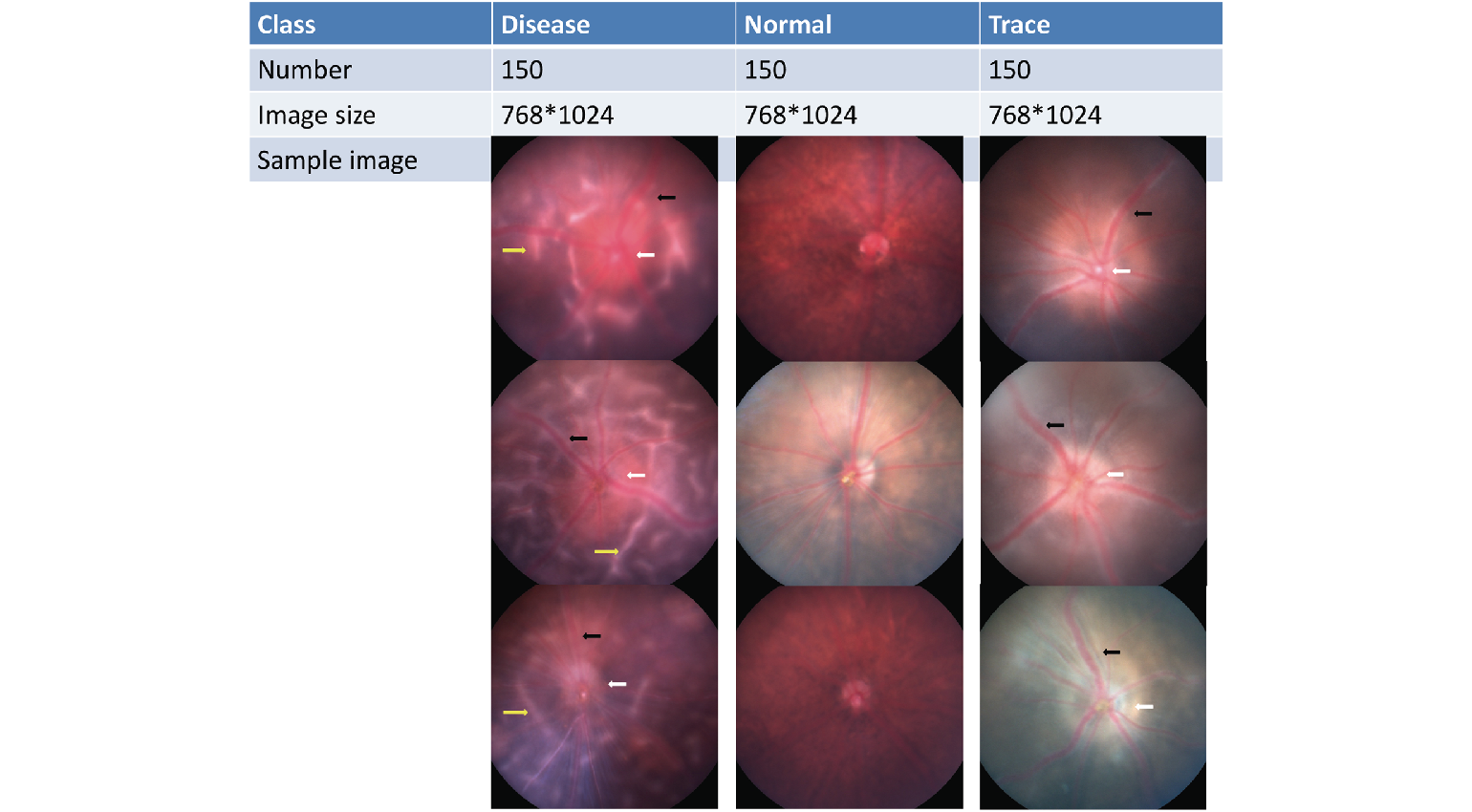
of 180 images and an external dataset of 33 images were used for additional testing. White arrow, optic disc edema; black arrow, vasculitis; yellow arrow, retina folds.
In their paper “Identifying Mouse Autoimmune Uveitis from Fundus Photographs Using Deep Learning” the authors suggest that although there is more work to do, the application of deep learning to experimental animal research to extract features from images can lead to standardization in the interpretation of results across different laboratories, with the potential for more accurate comparisons and collaborations. Their paper also details how they built, trained and validated their CNN to provide an unbiased characterization of uveitis in mice based on the analysis of 1500 Phoenix Micron fundus images.
Citation: Sun J, Huang X, Egwuagu C, Badr Y, Dryden SC, Fowler BT, Yousefi S. Identifying mouse autoimmune uveitis from fundus photographs using deep learning. Trans Vis Sci Tech. 2020;9(2):59. You can read their paper here.